In my previous post, I followed up on my k-means tutorial by applying it to cluster MNIST. We’re used to reading digits, so the centroids made perfect sense to us since plotting them just looked like a smudged number. The dataset even clustered pretty well since all digits are centred and look similar enough that decisions can directly be made on individual pixels. However, if we imagine those 784 pixels to be sensor readings, it would likely be really hard to make sense of them. Additionally, many situations would benefit from some nonlinear transformations to better capture the relationships between features. What we want, therefore, is to reduce the original dimensionality, so clustering becomes more manageable.
In this post, we’ll try to pass the data through a neural network which outputs in three dimensions. We then use K-means directly as a loss function, trying to get it to cluster points close to their assigned centroids and far from the others. This method is supposed to fail. However, I hope to use this as an entry point to some more interesting methods. Essentially, I also just thought I’d show it off since I haven’t seen it presented before.
Build the model
We do the preliminaries and create our model. I made a model with two CNN layers and MaxPooling into a 3 unit dense output. In order to plot it in 3 dimensions, I chose 3. However, I tried it with 10 as well and it didn’t perform any better.
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
import random
from mpl_toolkits.mplot3d import Axes3D
import matplotlib.patches as mpatches
import math
colours = ['#00FA9A','#FFFF00','#2F4F4F','#8B0000','#FF4500','#2E8B57','#6A5ACD','#FF00FF','#A9A9A9','#0000FF']
# we want to split into 10 clusters, one for each digit
clusters_n = 10
# load MNIST data
(x_train, y_train), (x_test, y_test) = tf.keras.datasets.mnist.load_data()
# scale between 0 and 1
X = tf.constant(x_train/255.0)
class MyModel(tf.keras.Model):
def __init__(self):
super(MyModel, self).__init__()
self.cnn_1 = tf.keras.layers.Conv2D(filters=32, kernel_size=(3,3), activation="relu")
self.mp_1 = tf.keras.layers.MaxPool2D(pool_size=2)
self.cnn_2 = tf.keras.layers.Conv2D(filters=16, kernel_size=(5,5), activation="relu")
self.mp_2 = tf.keras.layers.MaxPool2D(pool_size=2)
self.flatten = tf.keras.layers.Flatten()
self.dense_1 = tf.keras.layers.Dense(12, activation="relu")
self.dense_2 = tf.keras.layers.Dense(3)
def call(self, x):
inner = self.cnn_1(x)
inner = self.mp_1(inner)
inner = self.cnn_2(inner)
inner = self.mp_2(inner)
inner = self.flatten(inner)
inner = self.dense_1(inner)
inner = self.dense_2(inner)
return inner
We need to pass the centroids as parameters both since we want to keep updating the same ones, and since GradientTape doesn’t like random initialisations anyways. The new function is half of the code for centroid updating, which we’ll use just to perform prediction once we have the model outputs and the trained centroids.
@tf.function
def update_centroids(orig_points, points_expanded, centroids):
centroids_expanded = tf.expand_dims(centroids, 1)
distances = tf.subtract(centroids_expanded, points_expanded)
distances = tf.square(distances)
distances = tf.reduce_sum(distances, 2)
assignments = tf.argmin(distances, 0)
means = []
for c in range(clusters_n):
eq_eq = tf.equal(assignments, c)
where_eq = tf.where(eq_eq)
ruc = tf.reshape(where_eq, [1,-1])
ruc = tf.gather(orig_points, ruc)
ruc = tf.reduce_mean(ruc, axis=[1])
means.append(ruc)
new_centroids = tf.concat(means, 0)
return new_centroids, assignments, distances
def do_kmeans(centroids, y_pred):
points_expanded = tf.expand_dims(y_pred, 0)
old_centroids = centroids
i = 0
while True and i < 50:
i+=1
centroids, assignments, distances = update_centroids(y_pred, points_expanded, centroids)
if tf.reduce_all(tf.equal(old_centroids, centroids)):
break
old_centroids = centroids
return centroids, assignments, distances
# classify points using trained centroids
# same code as for update_centroids, but only returns the argmin
def get_assignments(centroids, y_pred):
points_expanded = tf.expand_dims(y_pred, 0)
centroids_expanded = tf.expand_dims(centroids, 1)
distances = tf.subtract(centroids_expanded, points_expanded)
distances = tf.square(distances)
distances = tf.reduce_sum(distances, 2)
assignments = tf.argmin(distances, 0)
return assignments
Training
The training loop is fairly standard. I batch the data so it fits in the GPU. I’m counting on the fact that 5000 digits are varied enough to not be detrimental to training, but I might be wrong.
EPOCHS = 1000
BATCH_SIZE = 5000
LR = 0.0001
# patience is the number of epochs until it stops training if no loss improvements
patience = 10
waited = 0
# initialise our model
model = MyModel()
# use adam as optimiser
adam = tf.keras.optimizers.Adam(learning_rate=LR)
# initialise a large initial loss on which to improve
prev_best_loss = 1e10
# variable to save prev best weights in
# we want to load it back after patience runs out
prev_weights = None
# define centroids which we'll keep updating
g_centroids = None
for e in range(EPOCHS):
# shuffle data each time
shuffled_data = tf.random.shuffle(X)
# we'll batch it so it fits in GPU memory
batched_data = tf.reshape(shuffled_data, (-1, BATCH_SIZE, 28, 28, 1))
print(f"Epoch {e+1}", end="")
# variable to keep track of total epoch loss
tot_epoch_loss = 0
for idx, batch in enumerate(batched_data):
with tf.GradientTape() as g:
# predict
output = model(batch)
# take first clusters_n outputs as initialisation for centroids
if g_centroids is None:
g_centroids = output[:clusters_n]
# now we do k-means on the output of the model
g_centroids, assignments, distances = do_kmeans(g_centroids, output)
# compute the sum of minimum distances to a centroid
# in other words, the sum of distances between all points and their assigned centroid
# we want to minimise this to get them as tight as possible
dis_to_c = tf.reduce_sum(tf.reduce_min(distances, 0))
# we then want to also maximise the distance from all points to all other centroids
# otherwise the model would clump everything together
# we can get this by summing all distances and then subtracting the smallest ones
dis_to_all = tf.reduce_sum(distances)
dis_to_others = dis_to_all - dis_to_c
loss = dis_to_c/float(BATCH_SIZE) + 1/(dis_to_others/float(BATCH_SIZE))
# I had the loss go to NaN before
# pretty sure I fixed it but I'm not risking it
if math.isnan(loss):
print("N", end="")
continue
tot_epoch_loss += loss
gradients = g.gradient(loss, model.trainable_variables)
print(f".", end="")
adam.apply_gradients(zip(gradients, model.variables))
print(f"Epoch loss {tot_epoch_loss:.2f} ", end="")
# if best loss save the weights and reset patience
if tot_epoch_loss < prev_best_loss:
prev_weights = model.get_weights()
prev_best_loss = tot_epoch_loss
waited = 0
else:
waited += 1
print(f"Patience {waited}/{patience}")
# if no more patience load best weights and quit
if waited >= patience:
model.set_weights(prev_weights)
break
Training takes a while, depending on your hardware.
Results
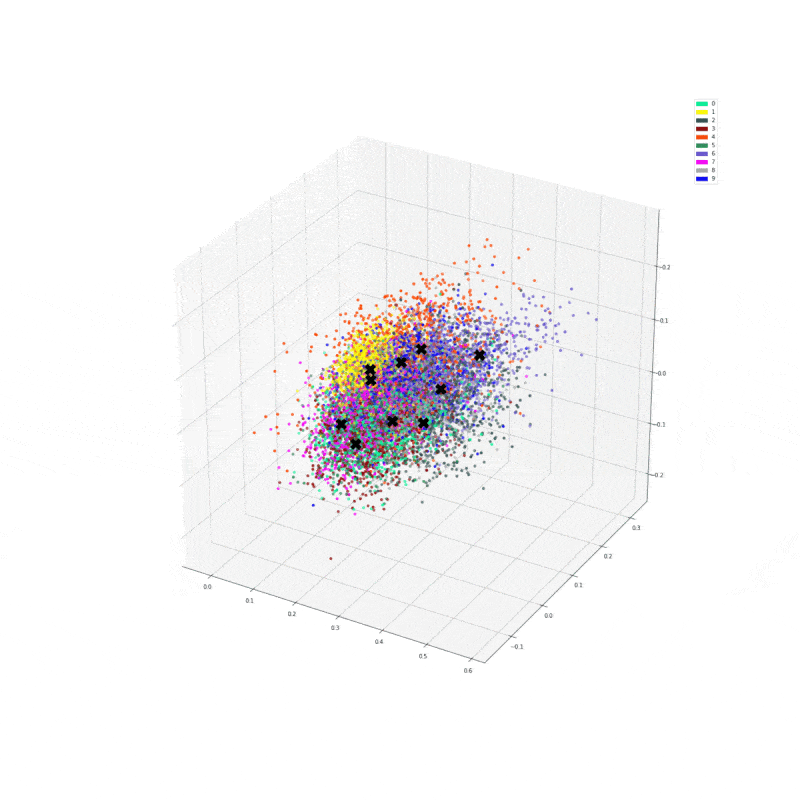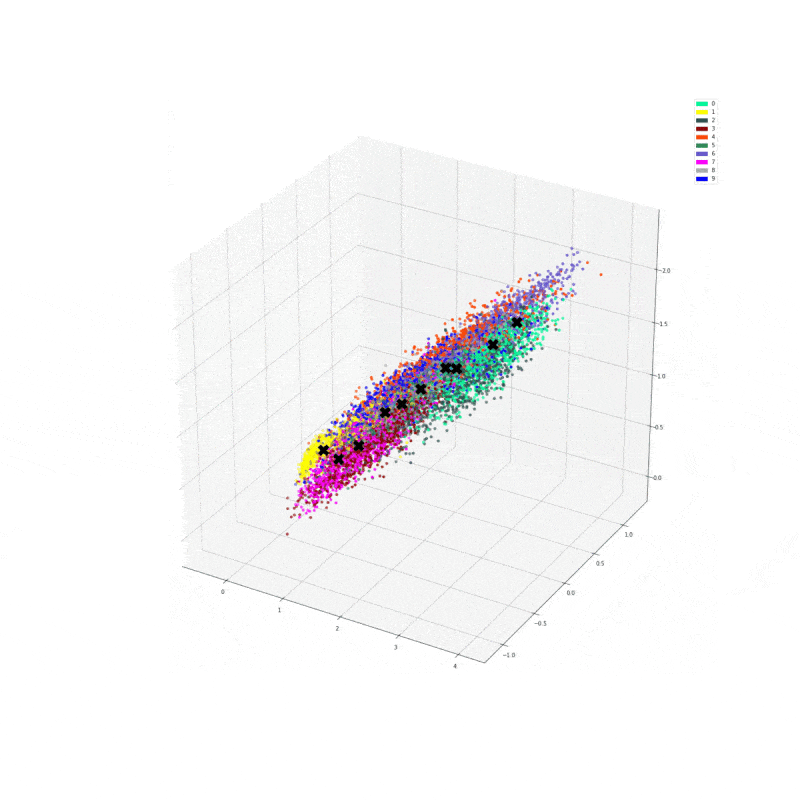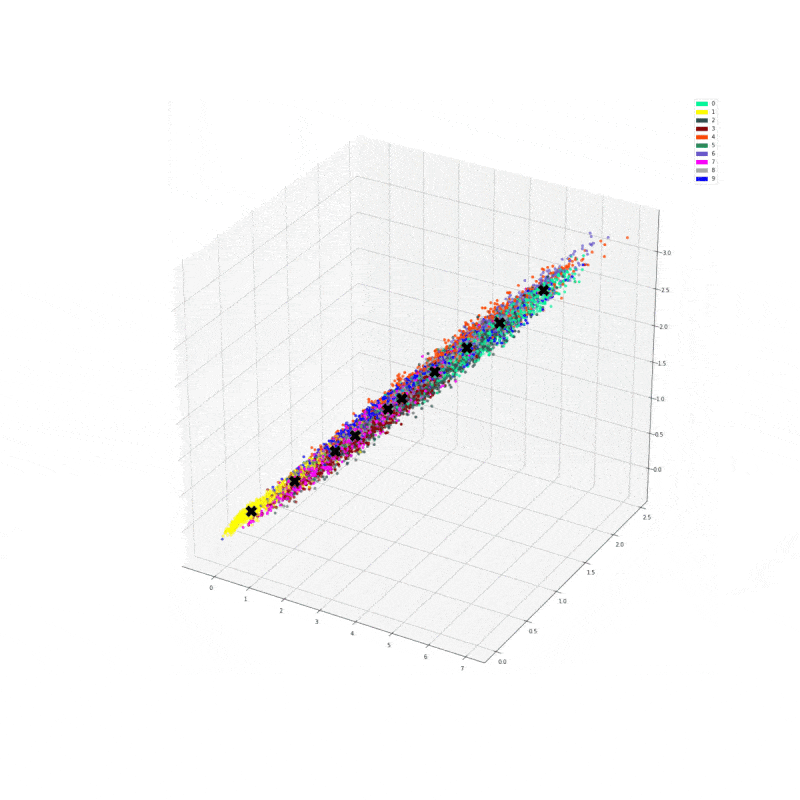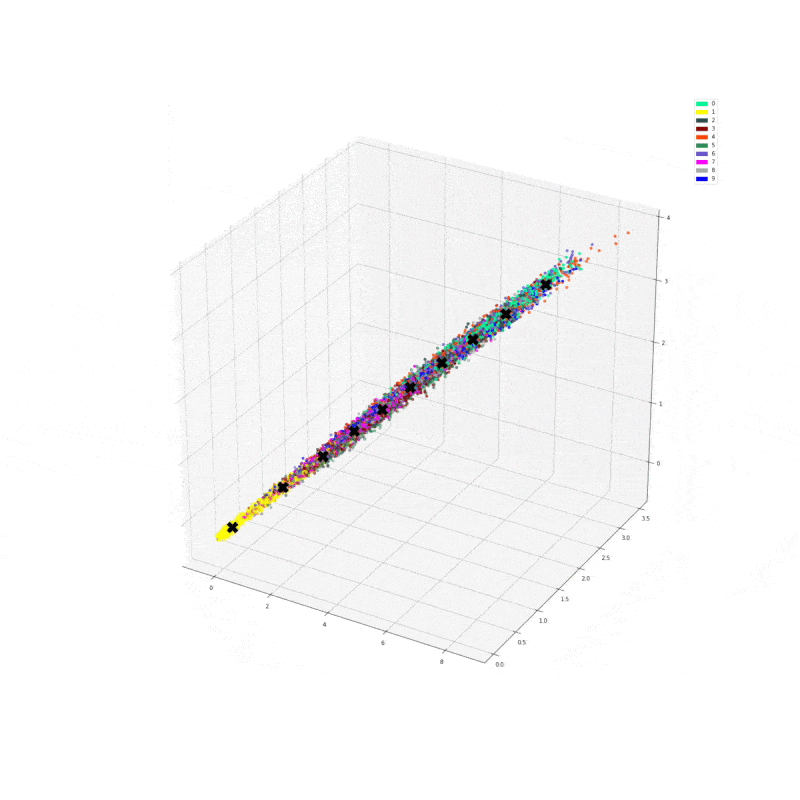
We can make a function that plots the numbers in the new feature space. We predict a number of centroids, scatter them with the corresponding colour, then plot the centroids.
def plot_dataset(X, y, model, centroids):
# Perform prediction on the dataset to get the intermediate representation
predict_batch_size = 10000
predict_count = 10000
m = []
for i in range(0, predict_count, predict_batch_size):
m.append(model(tf.reshape(X[i:i+predict_batch_size], (-1, 28, 28, 1))))
res = tf.concat(m, 0)
# scatter the points in the embedded feature space and the centroids
fig = plt.figure(figsize=(20,20))
ax = fig.add_subplot(111, projection='3d')
ax.scatter(res[:,0], res[:,1], res[:,2], zorder=1, color=[colours[y] for y in y_train[:predict_count]])
ax.plot(centroids[:, 0], centroids[:, 1], centroids[:, 2], "kX", markersize=20, zorder=1000)
# The following just handles printing the colours in the legend
mpc = []
for i in range(10):
mpatch = mpatches.Patch(color=colours[i], label=i)
mpc.append(mpatch)
plt.legend(handles=mpc)
plot_dataset(X, y_train, model, g_centroids)
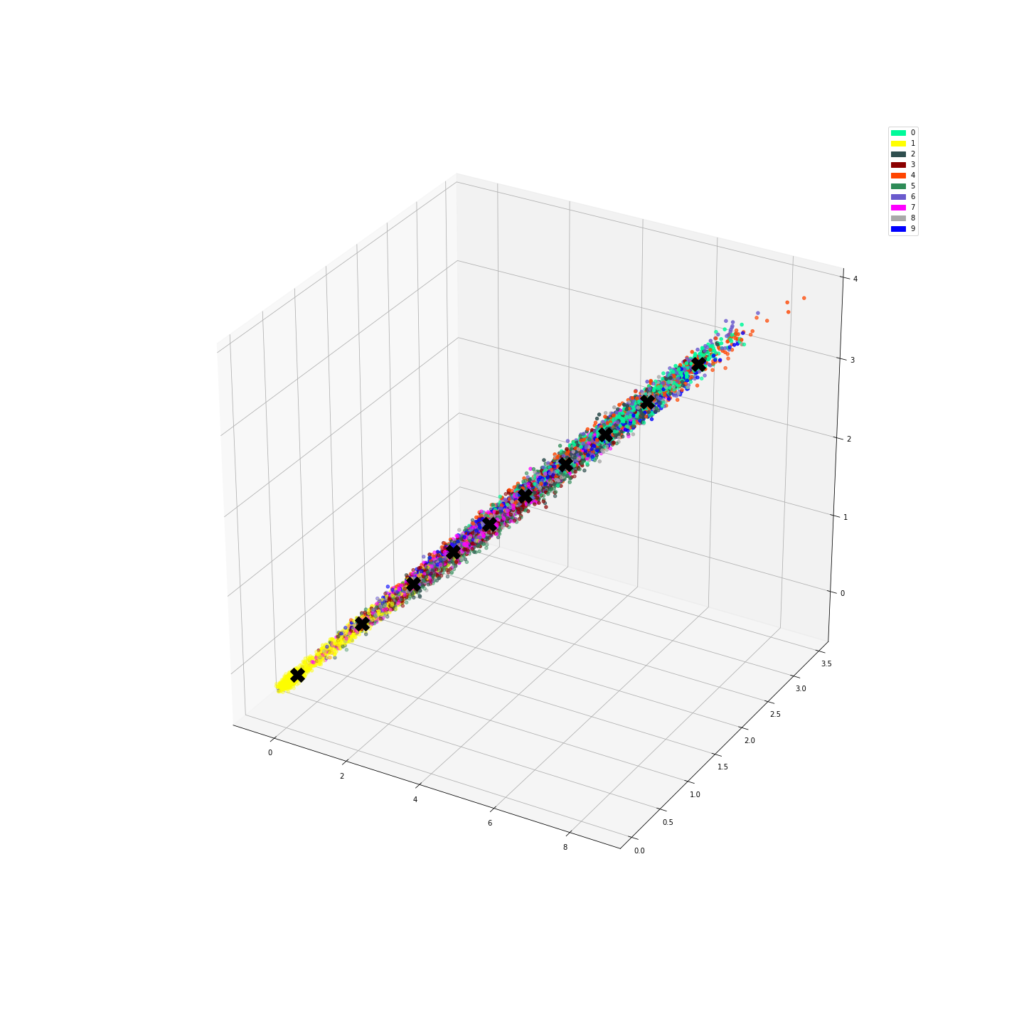
It seems it did something and managed to somehow cluster a few of the digits together. Finally, let’s calculate purity.
def calc_purity(labels, assignments):
d = np.zeros((clusters_n, clusters_n), dtype="int32")
for l, a in zip(labels, assignments):
d[a][l] += 1
purity_per_class = d.max(1)/d.sum(1)
# some are NaN
purity_per_class = purity_per_class[~np.isnan(purity_per_class)]
return np.mean(purity_per_class)
assignments = get_assignments(g_centroids, res)
calc_purity(y_train, assignments)
0.3153489094910568
Conclusion
It’s much worse than last time. Oh well. I imagine it’s part of the reason why nobody uses this method.
In any case, I hope this at least showed an uncommon way to reduce dimensionality for clustering by using k-means directly as a loss function. Stick around, since I hope to be able to illustrate some better ways of doing this in the following few weeks. As usual, please let me know if you find any bugs in my code or suggestions for improvements.